paired end sequencing read length
The library prep protocols are designed to. Depending on your sequencing data.

Comparison Of Next Generation Sequencing Systems
1b with the highest contig N50 2454 Mb and the longest contig 7968 Mb.
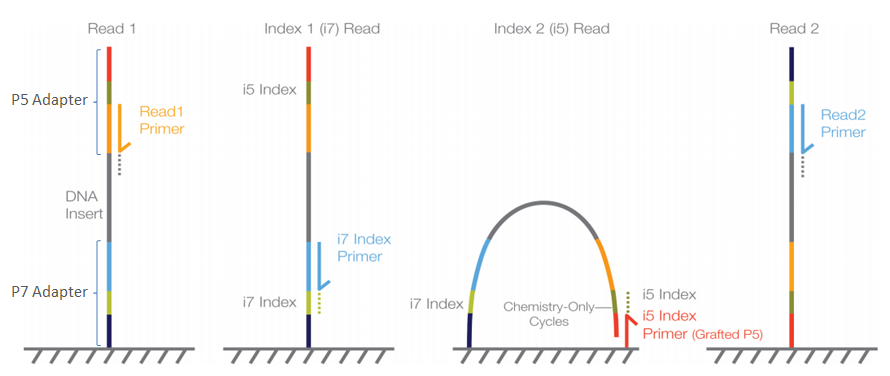
. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. The current read length that is standard for many experiments is paired-end 100 bp reads and there is also the possibility of running paired-end 300 bp reads. To merge paired reads select one or more sequence list documents and go to the Set merge paired reads option in the Pre-processing dropdown.
As long as you dont exceed the maximum number of cycles you will be fine. Contig length metrics were positively correlated with both read length and sequence coverage Fig. Many platforms Illumina Genome Analyzer Applied Biosystems.
We use an Illumina MiniSeq for our short-read sequencing runs. For Illumina kits for example you include R1 and R2 length in the sample sheet. Starting from micca 160 staggered read pairs staggered pairs are pairs where the 3 end of the reverse read has an overhang to the left of the 5 end of the forward read will be merged by.
In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. To ensure sequencing quality of the Index Read do not exceed the supported read. Next-generation sequencing technology is enabling massive production of high-quality paired-end reads.
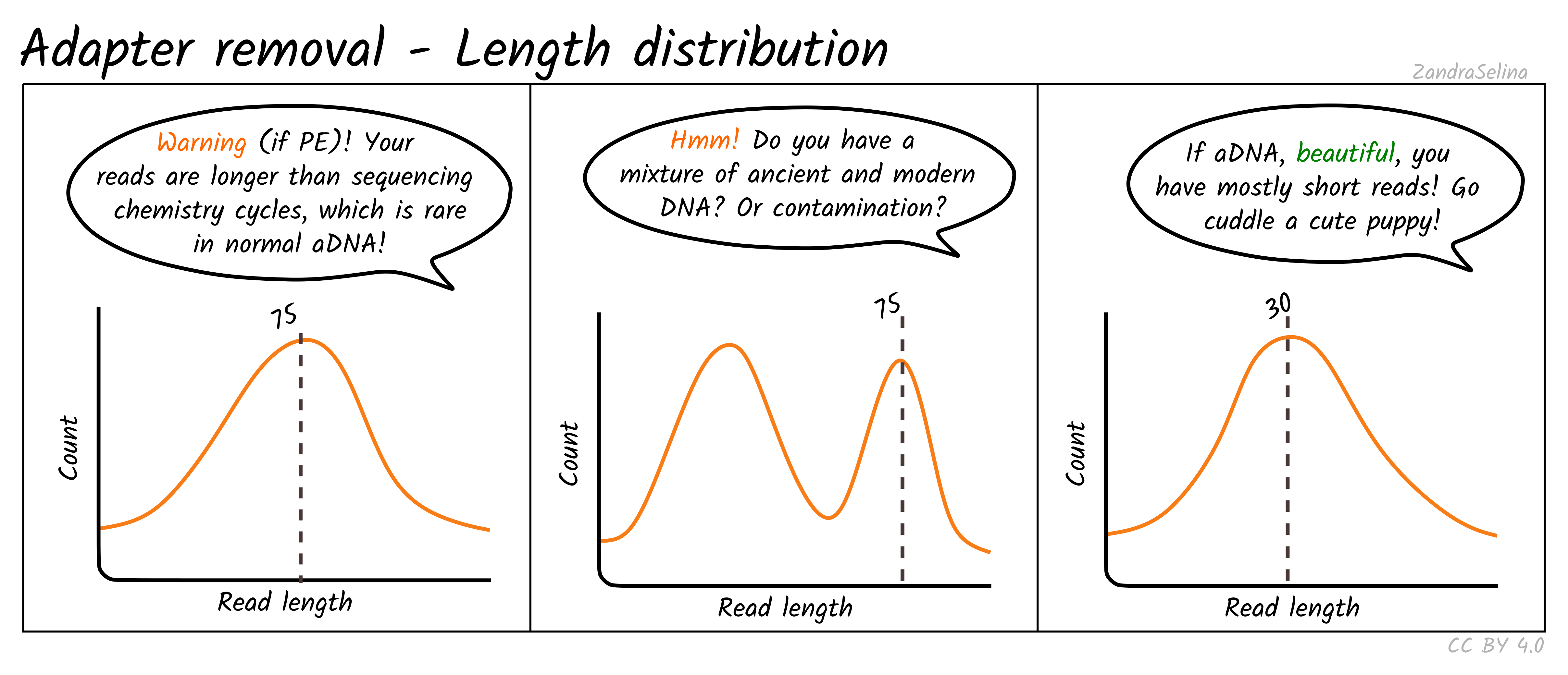
It is shown that re-sequencing and de novo sequencing of the majority of a bacterial genome is possible with read lengths of 2030 nt and that reads of 50 nt can provide. When a DNA fragment is shorter than two times the read length the paired reads overlap and can be merged into a longer read. For a 150 cycle kit.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Ideally merged reads can reach almost two. Modern nextgeneration sequencing platforms offer a range of read configurations such as single-read SR and paired-end PE sequencing with 75 bp per read 100 bp per read and 150 bp per.
The paired-end short read lengths are always 2 x 150bp 300bp. Paired-end sequencing facilitates detection of genomic. Enter up to 20 characters or use manual mode if you need between 20 and 100 bp.
Paired-end sequencing facilitates detection of genomic.

Schematic Representation Of Paired End Sequencing For Very Short Download Scientific Diagram
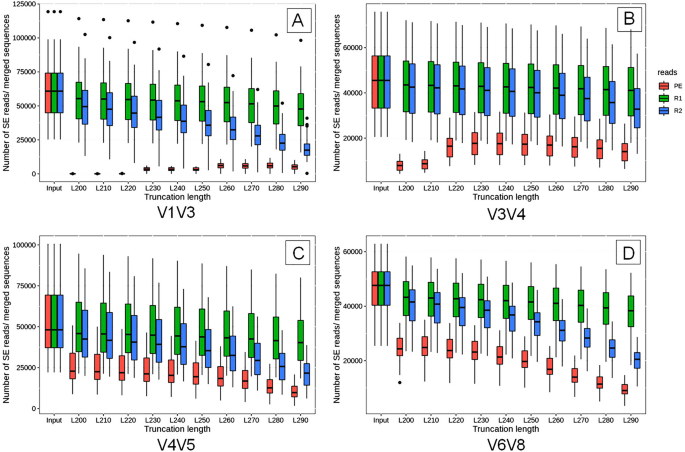
A Comprehensive Evaluation Of Single End Sequencing Data Analyses For Environmental Microbiome Research Springerlink
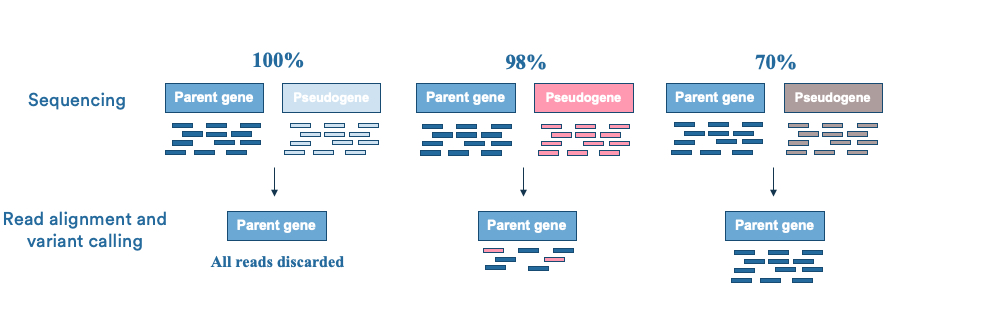
Blueprint Genetics Approach To Pseudogenes And Other Duplicated Genomic Regions Blueprint Genetics
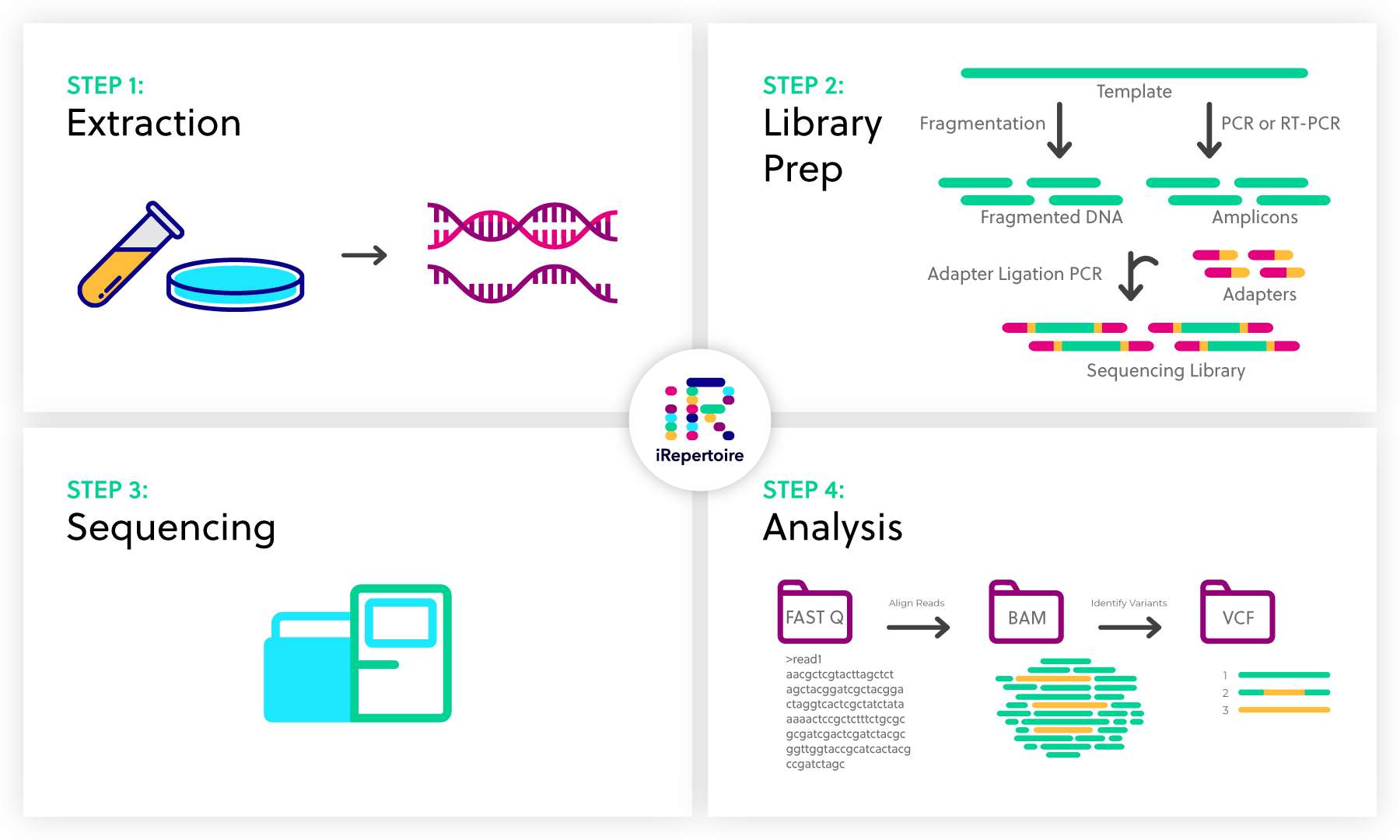
Next Generation Sequencing Ngs Overview Irepertoire Inc

How To Calculate The Coverage For A Ngs Experiment

What Is Mate Pair Sequencing For
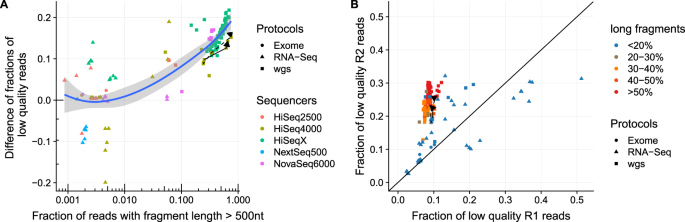
Long Fragments Achieve Lower Base Quality In Illumina Paired End Sequencing Scientific Reports

Next Generation Sequencing Technology Advances And Applications Sciencedirect
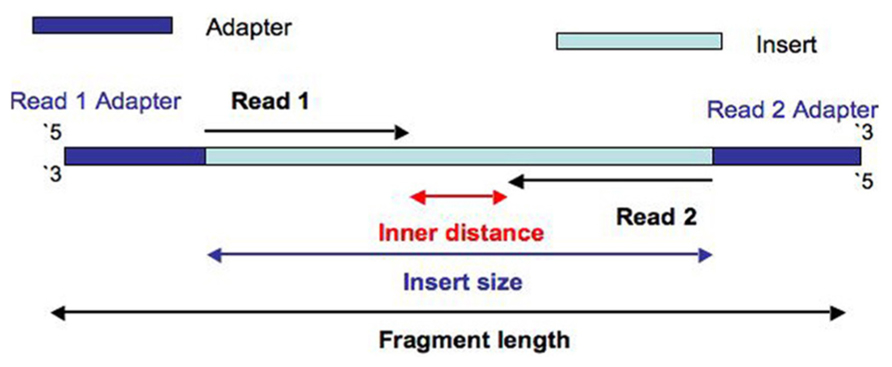
Why Do Illumina Reads All Have The Same Length When Sequencing Differently Sized Fragments

Illumina And Pacbio Dna Sequencing Data De Novo Assembly And Annotation Of The Genome Of Aurantiochytrium Limacinum Strain Ccap 4062 1 Sciencedirect

Beyond The Linear Genome Paired End Sequencing As A Biophysical Tool Trends In Cell Biology

Skewer A Fast And Accurate Adapter Trimmer For Next Generation Sequencing Paired End Reads Topic Of Research Paper In Biological Sciences Download Scholarly Article Pdf And Read For Free On Cyberleninka Open Science

Sequencing Read Length How To Calculate Ngs Read Length
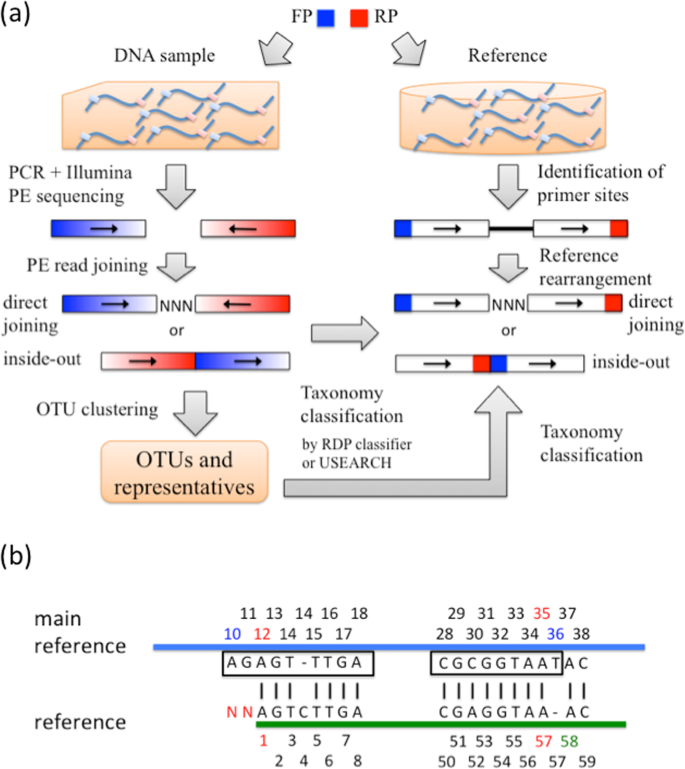
Joining Illumina Paired End Reads For Classifying Phylogenetic Marker Sequences Bmc Bioinformatics Full Text

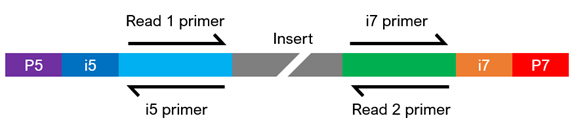
Paired End Sequencing Left Showing Read 1 And Read 2 Primers Starting Download Scientific Diagram